

MAFFT version 7
wsl --installto install Ubuntu.
wslto start Ubuntu.
Details: https://docs.microsoft.com/windows/wsl/install
$ wget https://mafft.cbrc.jp/alignment/software/mafft_7.526-1_amd64.deb
To paste this command into the Ubuntu window, use right button of mouse.
If it does not work, enable QuickEdit Mode.
$ sudo dpkg -i mafft_7.526-1_amd64.deb [sudo] password for username: (Type the password that was set in step 2)
$ which mafft /usr/bin/mafft $ mafft --version v7.526 (2024/Apr/22)
In short, C: on Windows = /mnt/c on Ubuntu.
$ cd /mnt/c/Users/yourname/DATA
To check if input.txt actually exists in this folder, use the ls command.
$ ls input.txt
$ mafftand follow the prompts to select input file, output file and options.
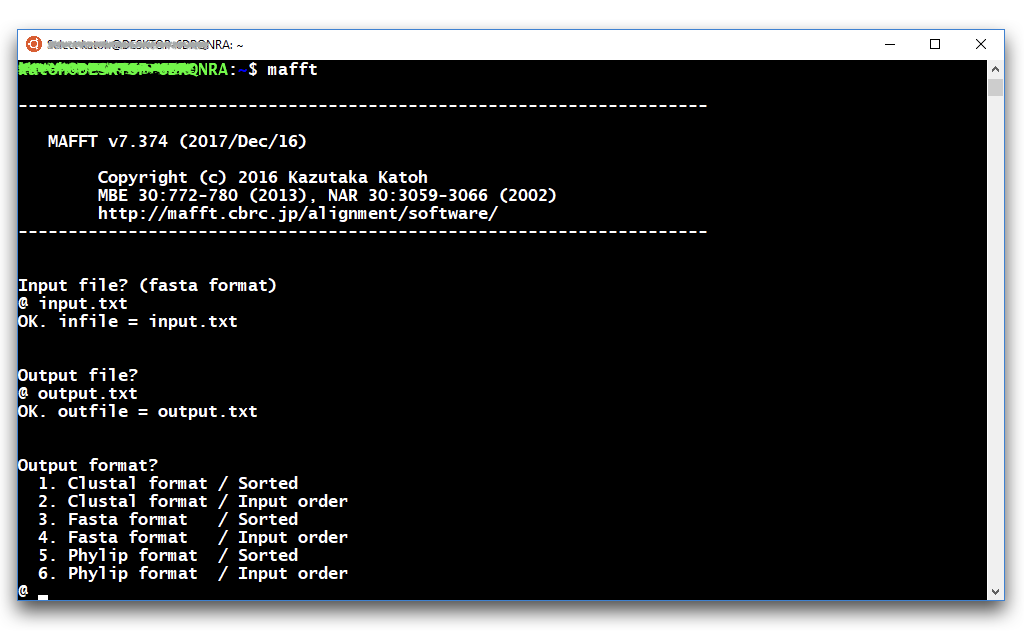
$ mafft (options) input.txt > output.txt
All features can be used in this mode.
C:\somewhere> wsl.exe mafft (options) --out output.txt input.txt
Not yet tested.
$ sudo apt install ruby
$ ruby -pe 'sub("\n","\r\n")' output.txt > output.windows.txt